Roadmap
info
Last update on 2021-02-03
GenoMLv1 started as a beta project that lasted for a year; it had >600 unique users and let the team get a lot of feedback from the community. It was an aggregation of code we have been using internally for quite some time, all of it coming from a variety of different projects and applications. Today, we've released a much-improved version of GenoML that will build a solid foundation for a modular extension. We are very excited about the GenoML expansion from just an AutoML to a broader framework. The new GenoML enables an ecosystem of machine learning tools for genomics.
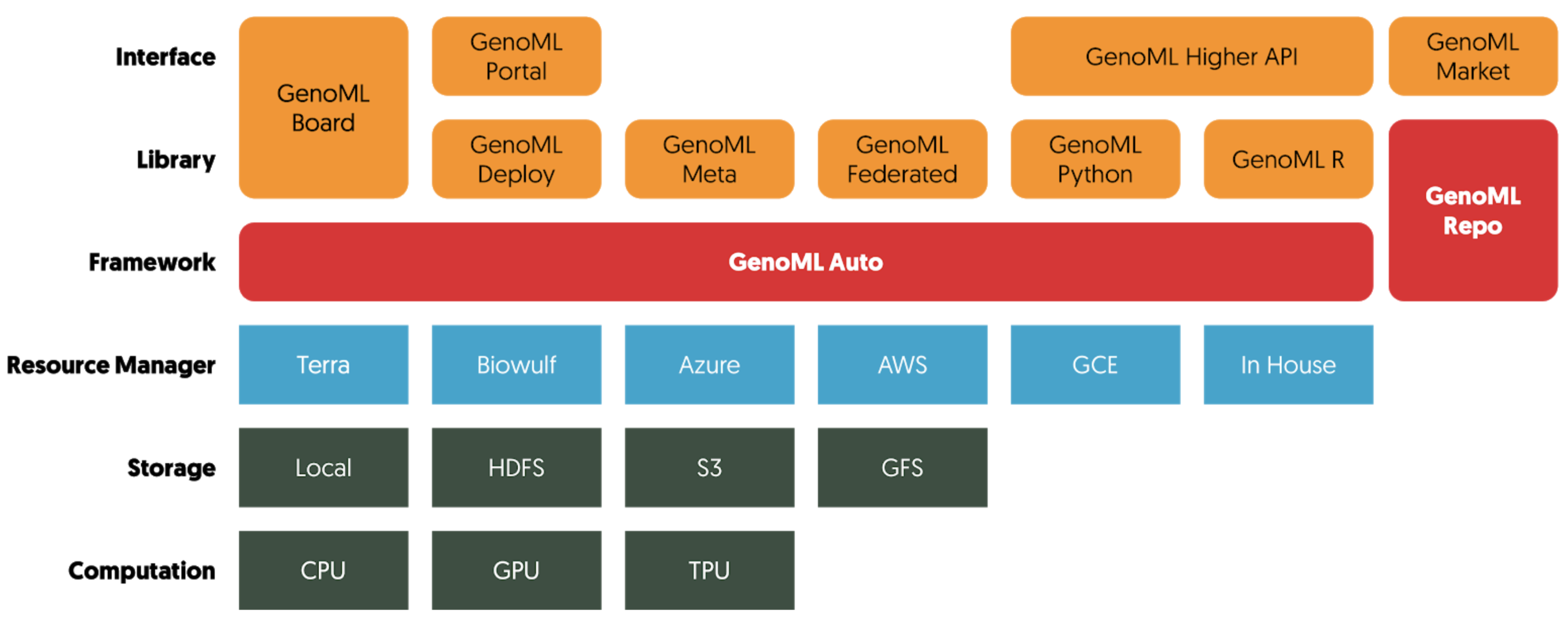
The framework layer, GenoML Auto, is the GenoMLv2 package. In the v3, it will be redesigned to enable the new upper layers. It will also be tested to be compatible with the “Resource manager” layer supporting a broad range of platforms. The core developer team maintains GenoML Auto. Contributions from the community are welcome.
The GenoML ecosystem components include:
- GenoML Genetics: general genetics pipeline tools
- GenoML Deploy: designed for deploying the ML models for inference in practice
- GenoML Portal: an interface designed for clinicians/physicians for use in practice. It also provides model explanation information
- GenoML Federated: federated learning of GenoML. A critical component in light of recent privacy regulations such as GDPR. Enables learning across multiple data silos
- GenoML Meta: meta-learning aspect of GenoML. Enabling learning and selection across diverse datasets and study populations
- GenoML Python: library developed to integrate seamlessly with other scientific Python libraries
- GenoML Higher API: higher-level APIs, enabling more community developments
- GenoML Repo: a repository of developed machine learning models, GWAS summary stats, etc.
- GenoML Market: a place to share models trained on public or private datasets (model zoo)
- GenoML Board: similar to the TensorFlow board, is an interface designed for researchers to learn, try, and evaluate GenoML easily
As part of use cases, we will also be including automated versions of the workflows used to support work at the Global Parkinson’s Genetics Program and the NIH Center for Alzheimer’s and Related Dementias.
We've set some goals below and a loose timeline:
2020-2021 (GenoMLv2)#
This development cycle includes:
- Transitioning to a 100% Python core
- Generally making the pipeline and docs more user friendly
- Upping speed and efficiency of the pipeline
- GWAS QC and Pipeline
- Integrating additional ML algorithm
- Network analyses
- Biobank-scale support
- Cross-silo checks for genetic duplicates
- Outlier detection
- Adding supervised prediction multi-class
- Adding unsupervised clustering multi-class
- Using models for production
2021-2022 (GenoMLv3)#
GenoML v3 is a lot of work, but it is crucial for the future of multi-omics global-scale research projects:
- GenoML Auto: adding support for longitudenal data
- GenoML Auto: Enhanced genetic mdoels for rare events and outliers
- GenoML Deploy
- GenoML Repo
- GenoML Meta
- GenoML Federated